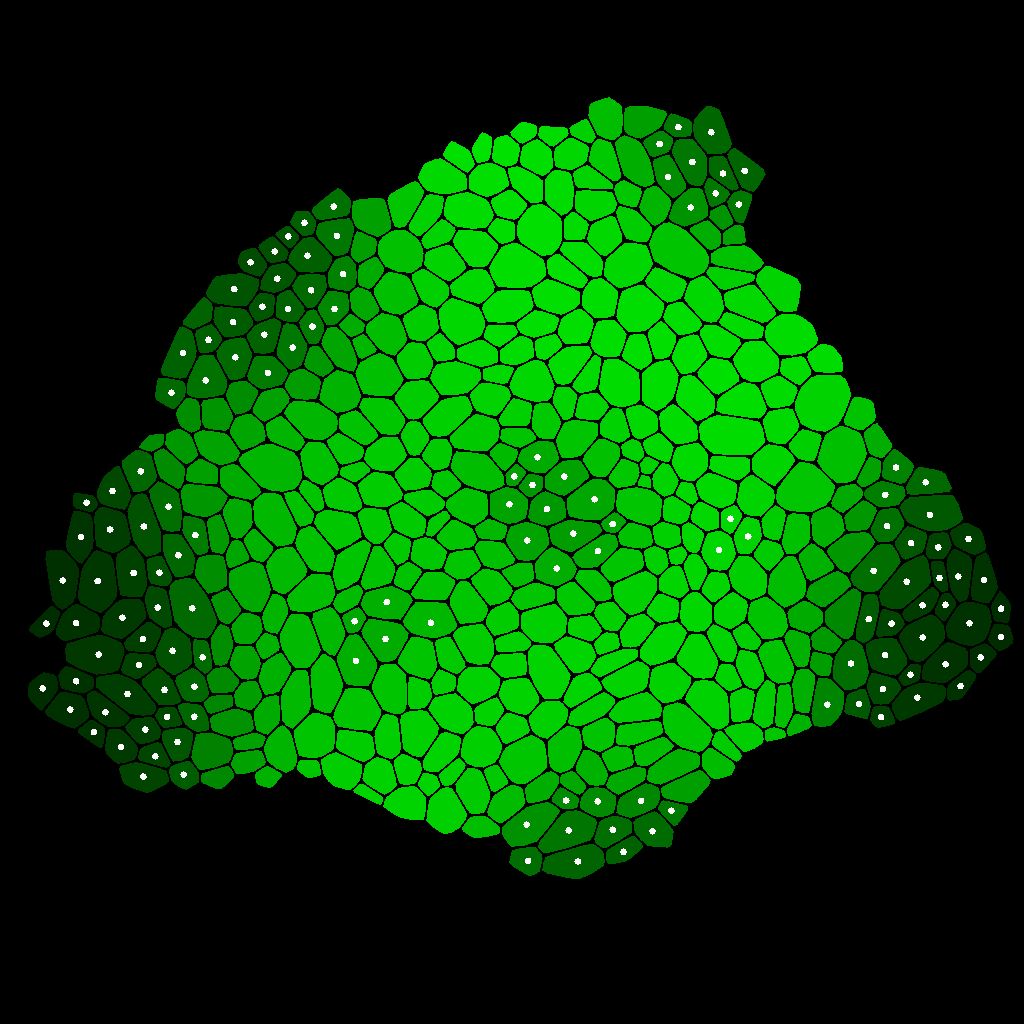
Simple Diffusion
This is an example showing how a simple physiological process can be simulated using STSE. Here, we consider the distribution of a morphogen A on a tissue acquired from .dat file. We assume that the morphogen A moves due to cell-cell diffusion, is produced everywhere and is degraded in the white dotted cells. The diffusion is simplified, which means this is rather qualitative then quantitative example. The visual result is presented at the bottom.
from openalea.stse.io.walled_tissue.dat_representation import read_dat2walled_tissue, read_link_file
import openalea.plantgl.all as pgl
import openalea.plantgl.ext.all as pd
from lib_09_06_06_influenceZones import vis, display_config
from openalea.stse.io.walled_tissue.dat_config_processing import read_dat_tissue_directory, Config
from openalea.stse.visu.walled_tissue_pgl import visualisation_pgl_2D_varried_membrane_thickness, f_property2scalar, f_cell_marking, f_properties2material, f_weighted_property2material
from openalea.stse.structures.algo.walled_tissue import wv_edge2cell_edge
import scipy
import scipy.integrate.odepack
import copy
class DiffusionAction:
"""Diffusion of a substance A in a WalledTissue.
"""
def __init__(self, tissue = None):
self.t = 0
self.h = 10
self.c_change = 0.0001 #acceptable change could not be smaller to classifie as error.
self.c_max_steps = 100 #maximum number of steps
self.local_evacuation_properties = ["P1","P2","P3","P4","P5","P6",]
self.c_alpha = 0.01 #creation
self.c_beta = 0.01 #destruction
self.c_gamma = 0.05#diffusion
self.c_kappa = 0.05 #diffusion
self.c_epsilon=0.01 #centre evacuate
self.c_delta = 0.01 #prim evacuate
self.c_initial_A_value=self.c_alpha/self.c_beta
self.tissue = tissue
self.init_A()
def A( self, cell, value=None ):
return self.tissue.cell_property( cell, "A", value=value)
def apply( self ):
self.stable_step( )
def stable_step( self ):
t = self.tissue
self.prepare_sys_map()
stable=False
i=0
while not stable and i < self.c_max_steps:
print " #: physiology A diffusion: loop=", i, " time=",self.t
i+=1
res= scipy.integrate.odepack.odeint( self.f, self.prepare_x0(), [self.t, self.t+self.h],rtol=0.01, atol=0.001 )
self.t += self.h
stable = self.rate_error( res[0], res[1])
self.update( res[1] )
def update( self, x ):
t = self.tissue
for i in t.cells():
t0 = x[ self.cell2sys[ i ] ]
t0 = max(0,t0)
self.A( cell=i, value= t0)
def rate_error( self, xn, xnprim ):
t = True
max = -float("infinity")
for i in range( len( self.tissue.cells() )):
if abs( xn[i] - xnprim[i] ) > max:
max=abs( xn[i] - xnprim[i] )
if self.c_change < abs( xn[i] - xnprim[i] ):
t=False
print " #: errror = ", max# abs( xn[i] - xnprim[i] )
return t
def prepare_x0( self ):
x0 = []
t = self.tissue
for x in range( len(t.cells()) ):
x0.append(0)
for i in t.cells():
x0[ self.cell2sys[ i ] ] = self.A( i )
return x0
def prepare_sys_map( self ):
self.cell2sys = {}
t = self.tissue
ind = 0
for i in t.cells():
self.cell2sys[ i ] = ind
ind += 1
def f( self, x, t ):
def D( (i, j), t, x):
return (A(i,t,x)-A(j,t,x))
def A( cid, t, x ):
return x[ self.cell2sys[ cid ] ]
t = self.tissue
x2 = copy.copy( x )
for i in t.cells():
x2[ self.cell2sys[ i ] ] = self.c_alpha - A(i,t,x)*(self.c_beta+self.c_epsilon*self.i_center_evacuate( i ))#OK
if self.i_local_evacuate( i ):
x2[ self.cell2sys[ i ] ] -= self.c_delta*A(i,t,x)
for n in t.cell_neighbors( cell=i ):
x2[ self.cell2sys[ i ] ] += self.c_gamma*D((n,i),t,x)
return x2
def i_local_evacuate( self, cell ):
t = self.tissue
for i in self.local_evacuation_properties:
if t.cell_property( cell=cell, property= i ):
return 1
return 0
def i_center_evacuate( self, cell ):
t = self.tissue
if t.cell_property( cell=cell, property="CZ") > 0:
return 1
else:
return 0
def init_A(self):
t = self.tissue
for c in t.cells():
self.A(cell= c, value= self.c_initial_A_value ) #+random.random() )
class TissueSystem:
"""Structure to store different components of tissue-based experiments.
"""
def __init__( self, wt=None, config=None, phys=[] ):
"""Inits tissue system
<Long description of the function functionality.>
:parameters:
arg1 : WalledTissue
Tissue structure
"""
self.tissue = wt
self.config = config
self.phys = phys
self.frame = 0
self.visualise()
def simulate( self, steps=1 ):
"""Simulates all physiological processes.
<Long description of the function functionality.>
"""
for j in range( steps ):
for i in self.phys:
i.apply()
self.visualise()
def visualise( self, save=False, clear=True ):
"""Simple visualisation using PlantGL. The colormap is linked to
morphogen gradient.
"""
if clear: pd.SCENES[ pd.CURRENT_SCENE ].clear()
pd.instant_update_viewer()
visualisation_pgl_2D_varried_membrane_thickness( self.tissue,
abs_intercellular_space=0.05,
abs_membrane_space=0.25,
stride=20,
#f_membrane_thickness = f_pin( self.tissue ),
f_cell_marking = [f_cell_marking( properties=self.config.cell_regions.keys(), property_true_radius=0.2)],
f_material = f_weighted_property2material( property="A", range=(0,1) ),
)
pd.instant_update_viewer()
self.frame += 1
if save: pgl.Viewer.frameGL.saveImage( config.file_folder+"/"+str(self.frame).zfill(5)+".png" )
path = "/Users/stymek/src/stse/trunk/data/09-06-10-marianne-wt2-diff/"
wt = read_dat_tissue_directory( path +"config.py" )
c = Config( path+"config.py" )
ts = TissueSystem( wt=wt, config=c, phys=[ DiffusionAction( tissue=wt ) ])
ts.simulate()
ts.visualise()
display_config()
import openalea.plantgl.all as pgl
import openalea.plantgl.ext.all as pd
from lib_09_06_06_influenceZones import vis, display_config
from openalea.stse.io.walled_tissue.dat_config_processing import read_dat_tissue_directory, Config
from openalea.stse.visu.walled_tissue_pgl import visualisation_pgl_2D_varried_membrane_thickness, f_property2scalar, f_cell_marking, f_properties2material, f_weighted_property2material
from openalea.stse.structures.algo.walled_tissue import wv_edge2cell_edge
import scipy
import scipy.integrate.odepack
import copy
class DiffusionAction:
"""Diffusion of a substance A in a WalledTissue.
"""
def __init__(self, tissue = None):
self.t = 0
self.h = 10
self.c_change = 0.0001 #acceptable change could not be smaller to classifie as error.
self.c_max_steps = 100 #maximum number of steps
self.local_evacuation_properties = ["P1","P2","P3","P4","P5","P6",]
self.c_alpha = 0.01 #creation
self.c_beta = 0.01 #destruction
self.c_gamma = 0.05#diffusion
self.c_kappa = 0.05 #diffusion
self.c_epsilon=0.01 #centre evacuate
self.c_delta = 0.01 #prim evacuate
self.c_initial_A_value=self.c_alpha/self.c_beta
self.tissue = tissue
self.init_A()
def A( self, cell, value=None ):
return self.tissue.cell_property( cell, "A", value=value)
def apply( self ):
self.stable_step( )
def stable_step( self ):
t = self.tissue
self.prepare_sys_map()
stable=False
i=0
while not stable and i < self.c_max_steps:
print " #: physiology A diffusion: loop=", i, " time=",self.t
i+=1
res= scipy.integrate.odepack.odeint( self.f, self.prepare_x0(), [self.t, self.t+self.h],rtol=0.01, atol=0.001 )
self.t += self.h
stable = self.rate_error( res[0], res[1])
self.update( res[1] )
def update( self, x ):
t = self.tissue
for i in t.cells():
t0 = x[ self.cell2sys[ i ] ]
t0 = max(0,t0)
self.A( cell=i, value= t0)
def rate_error( self, xn, xnprim ):
t = True
max = -float("infinity")
for i in range( len( self.tissue.cells() )):
if abs( xn[i] - xnprim[i] ) > max:
max=abs( xn[i] - xnprim[i] )
if self.c_change < abs( xn[i] - xnprim[i] ):
t=False
print " #: errror = ", max# abs( xn[i] - xnprim[i] )
return t
def prepare_x0( self ):
x0 = []
t = self.tissue
for x in range( len(t.cells()) ):
x0.append(0)
for i in t.cells():
x0[ self.cell2sys[ i ] ] = self.A( i )
return x0
def prepare_sys_map( self ):
self.cell2sys = {}
t = self.tissue
ind = 0
for i in t.cells():
self.cell2sys[ i ] = ind
ind += 1
def f( self, x, t ):
def D( (i, j), t, x):
return (A(i,t,x)-A(j,t,x))
def A( cid, t, x ):
return x[ self.cell2sys[ cid ] ]
t = self.tissue
x2 = copy.copy( x )
for i in t.cells():
x2[ self.cell2sys[ i ] ] = self.c_alpha - A(i,t,x)*(self.c_beta+self.c_epsilon*self.i_center_evacuate( i ))#OK
if self.i_local_evacuate( i ):
x2[ self.cell2sys[ i ] ] -= self.c_delta*A(i,t,x)
for n in t.cell_neighbors( cell=i ):
x2[ self.cell2sys[ i ] ] += self.c_gamma*D((n,i),t,x)
return x2
def i_local_evacuate( self, cell ):
t = self.tissue
for i in self.local_evacuation_properties:
if t.cell_property( cell=cell, property= i ):
return 1
return 0
def i_center_evacuate( self, cell ):
t = self.tissue
if t.cell_property( cell=cell, property="CZ") > 0:
return 1
else:
return 0
def init_A(self):
t = self.tissue
for c in t.cells():
self.A(cell= c, value= self.c_initial_A_value ) #+random.random() )
class TissueSystem:
"""Structure to store different components of tissue-based experiments.
"""
def __init__( self, wt=None, config=None, phys=[] ):
"""Inits tissue system
<Long description of the function functionality.>
:parameters:
arg1 : WalledTissue
Tissue structure
"""
self.tissue = wt
self.config = config
self.phys = phys
self.frame = 0
self.visualise()
def simulate( self, steps=1 ):
"""Simulates all physiological processes.
<Long description of the function functionality.>
"""
for j in range( steps ):
for i in self.phys:
i.apply()
self.visualise()
def visualise( self, save=False, clear=True ):
"""Simple visualisation using PlantGL. The colormap is linked to
morphogen gradient.
"""
if clear: pd.SCENES[ pd.CURRENT_SCENE ].clear()
pd.instant_update_viewer()
visualisation_pgl_2D_varried_membrane_thickness( self.tissue,
abs_intercellular_space=0.05,
abs_membrane_space=0.25,
stride=20,
#f_membrane_thickness = f_pin( self.tissue ),
f_cell_marking = [f_cell_marking( properties=self.config.cell_regions.keys(), property_true_radius=0.2)],
f_material = f_weighted_property2material( property="A", range=(0,1) ),
)
pd.instant_update_viewer()
self.frame += 1
if save: pgl.Viewer.frameGL.saveImage( config.file_folder+"/"+str(self.frame).zfill(5)+".png" )
path = "/Users/stymek/src/stse/trunk/data/09-06-10-marianne-wt2-diff/"
wt = read_dat_tissue_directory( path +"config.py" )
c = Config( path+"config.py" )
ts = TissueSystem( wt=wt, config=c, phys=[ DiffusionAction( tissue=wt ) ])
ts.simulate()
ts.visualise()
display_config()
Here is the result of the previous code: