Initialization
Here we present how to initialize the STSE structure, WalledTissue, with data. The simplest initialization can be done in the following way (please look at 09_05_01_wt_tutorial_01.py in the example folder).
from openalea.stse.structures.walled_tissue_const import WalledTissueConst
from openalea.stse.structures.algo.walled_tissue import create
from openalea.plantgl.math import Vector3
# we define the set of settings for a tissue
wtc = WalledTissueConst( name = "Tissue01")
# we declare the set of properties for every cell i.e.
# every cell will have P1 property which will be initialized
# by default to 0. the cell properties can be reached by:
# wt.cell_property( cell_id ) where wt is an instance of
# WalledTissue
# Similar properties can be defined for wv, wv_edges, cell_edges.
wtc.cell_properties = { "P1" : 0. }
# we create an empty tissue
wt = WalledTissue( const = wtc )
# we initialize the tissue by specifing two cells
# Please look at: help(create)
# Note: to understand this example geometry it is recomended to draw the cells
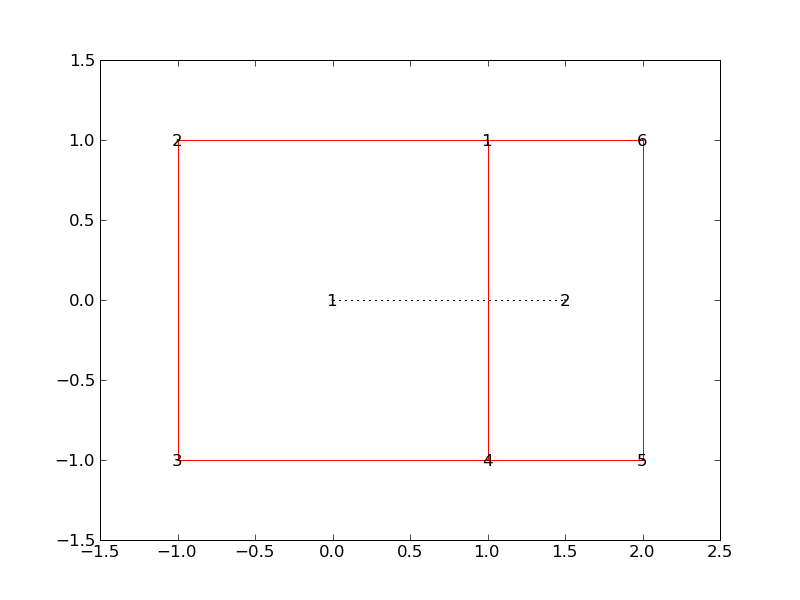
# first we specify cell corners and their geometry using the following mapping
wv2pos={
1: Vector3(1., 1., 0),
2: Vector3(-1., 1., 0),
3: Vector3(-1., -1., 0),
4: Vector3(1., -1., 0),
5: Vector3(2., -1., 0),
6: Vector3(2., 1., 0),
}
# second we specify cell shape of all cells
# it is important to note that the lists of vertices are ordered
cell2wv_list={
1: [1,2,3,4],
2: [1,4,5,6],
}
create(wt, wv2pos=wv2pos, cell2wv_list=cell2wv_list)
## sample cell inspection
from openalea.stse.structures.algo.walled_tissue import investigate_cell
investigate_cell(wt, 1)
## sample visualization using pylab
from openalea.stse.visu.walled_tissue_pylab import show_cells_with_wvs
show_cells_with_wvs(wt, True)
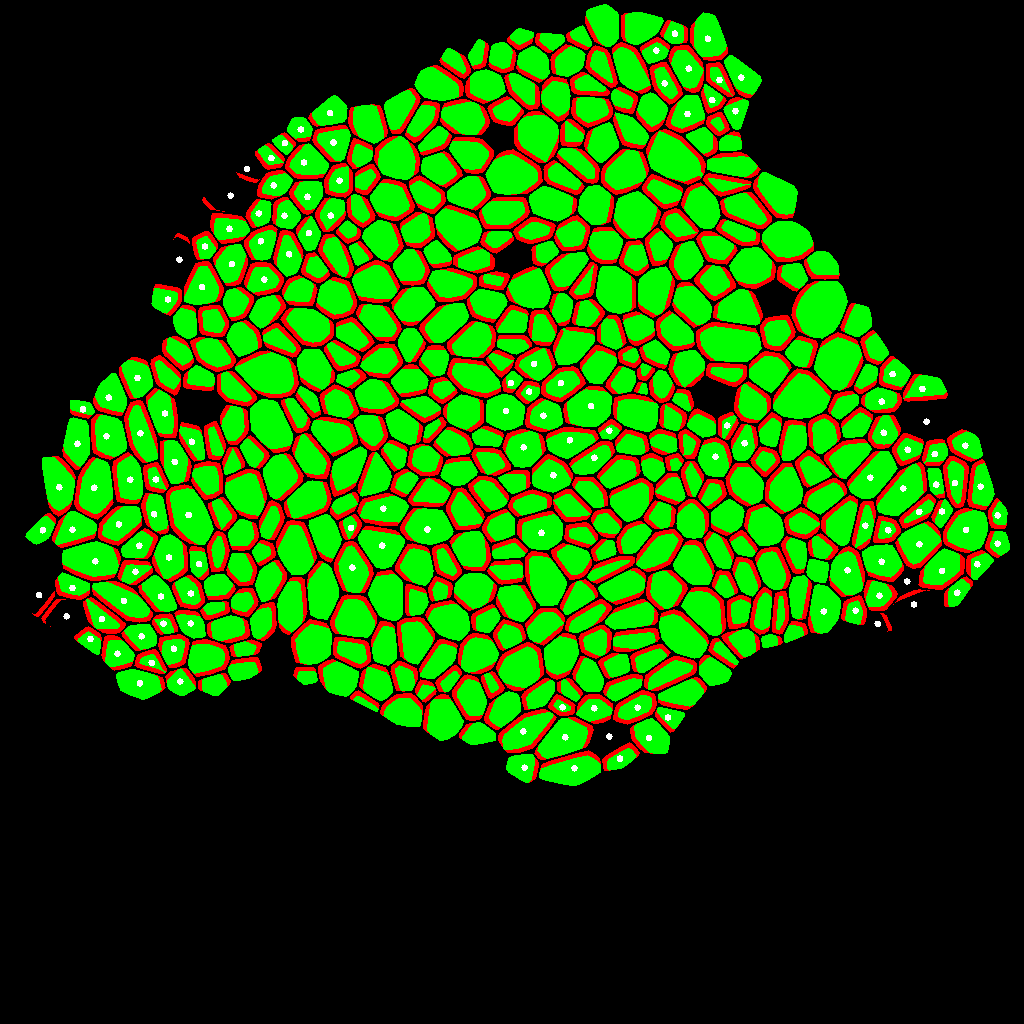
The result of the previous code displayed using pylab rutines:
Another way to initialize tissue might be using .dat file format. These files can be edited using merrydig software. The example of such an initialization can be found here (please look at 09_05_01_wt_tutorial_02.py in the example folder).
from openalea.stse.structures.walled_tissue_const import WalledTissueConst
from openalea.stse.structures.algo.walled_tissue import create
from openalea.stse.io.walled_tissue.dat_representation import read_dat2walled_tissue, read_link_file
from openalea.plantgl.math import Vector3
# we define the set of settings for a tissue
wtc = WalledTissueConst( name = "Tissue01")
# we declare the set of properties for every cell edge
# it corresponds to the protein PIN level
wtc.cell_edges_properties = { "PIN" : 0. }
# we create an empty tissue
wt = WalledTissue( const = wtc )
# please adjust the path to access the files from data directory of stsf
# project
stse_path = "/Users/stymek/src/stse/trunk/data"
# we initialize the tissue using the data read from
# .dat file Specification can be found in the stsf
# documentation
wt = read_dat2walled_tissue( filename = stse_path+"/2cellTest/2cellTest.dat", tissue_properties = wtc)
# setting physiological information
# read links from .link file
# .link file Specification can be found in the stse
# documentation
links = read_link_file( link_fn = stse_path+"/2cellTest/2cellTest.link")
for (i,j) in links:
try:
wt.directed_cell_edge_property((int(i), int(j)), "PIN", 1. )
except Exception:
print " ! link not added:", i,j
## adjusting visulalization
# adjusting the cell geometry to the display
avg=Vector3()
for i in wt.wvs():
wt.wv_pos(i, wt.wv_pos(i)*0.1)
avg+=wt.wv_pos(i)
avg = avg/float(len(wt.wvs()))
for i in wt.wvs():
wt.wv_pos(i, wt.wv_pos(i)-avg)
## sample visualization using plantGL
from openalea.stse.visu.walled_tissue_pgl import visualisation_pgl_2D_varried_membrane_thickness
visualisation_pgl_2D_varried_membrane_thickness( wt,
abs_intercellular_space=0.05,
abs_membrane_space=0.25
)
import openalea.plantgl.all as pgl
import openalea.plantgl.ext.all as pd
# viewer configuration
pgl.Viewer.camera.setOrthographic()
pgl.Viewer.display( pd.SCENES[ pd.CURRENT_SCENE ] )
pgl.Viewer.frameGL.setSize(1024,1024)
pgl.Viewer.camera.position = pgl.Vector3(0,0,30.)
pgl.Viewer.light.enabled=False
pd.instant_update_viewer()
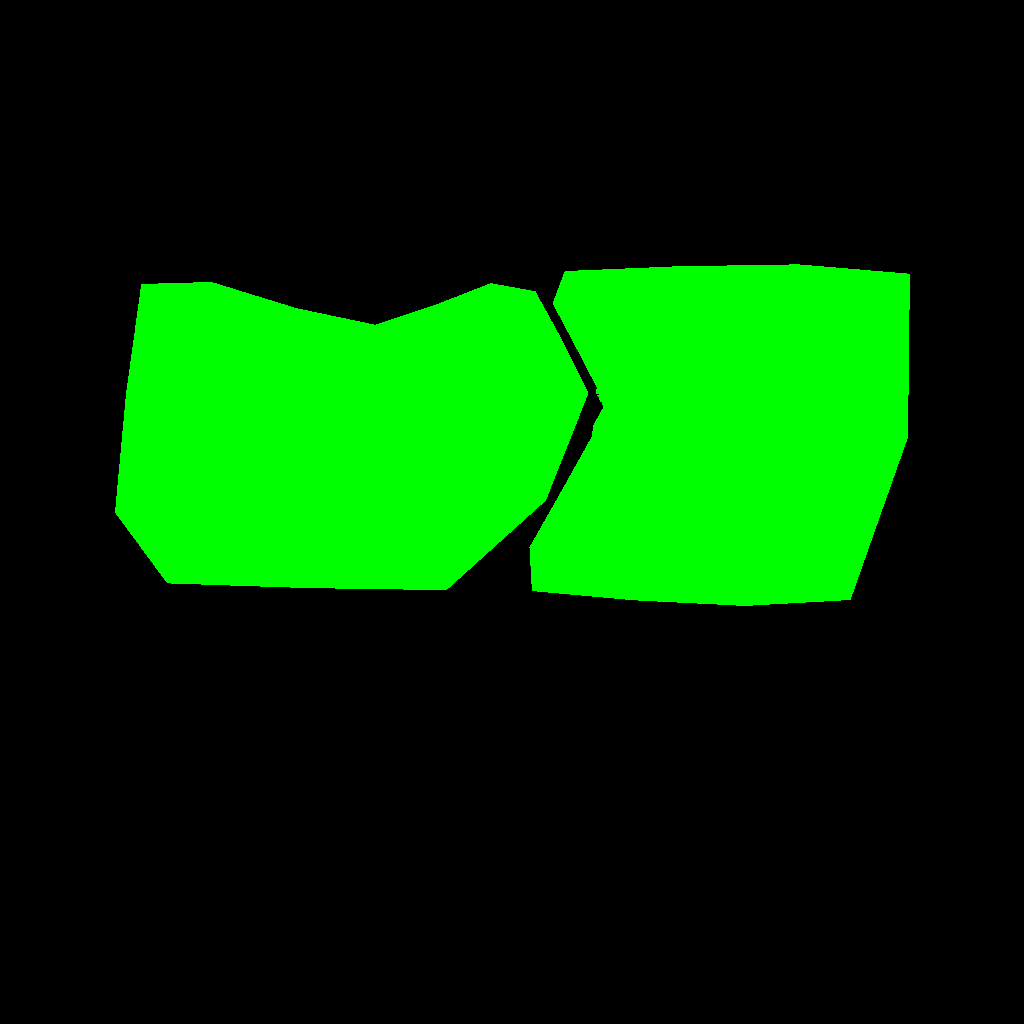
The result of such an initialization, printed using PlantGL rutines can look like this:
Another initalization can be done using the scripts facilitating the tissue management:
path = "/Users/stymek/src/stse/trunk/data/09-06-10-marianne-wt2/"
wt = read_dat_tissue_directory( path +"config.py" )
Then the config.py stored in tissue should look in a similar way:
# making sure that the configuration is new
import openalea.stse.io.walled_tissue.dat_config
m = reload( openalea.stse.io.walled_tissue.dat_config )
locals().update(m.__dict__)
format_version = 1.0
# scale use to convert the units
scale_factor = 0.1
# rotate the points around 0,0 and z-axis
rotation = math.pi/2.
# translate the points to make 0,0 geometric center
translation = True
#cells to remove
remove_cell_list = []
# path to data
file_folder = "/Users/stymek/src/stse/trunk/data/09-06-10-marianne-wt2"
# .dat file name
dat_file = "wt2.dat"
# .link file name
link_file = "wt2.link"
# link description in properties
link = "PIN"
#tissue description
description="Tissue of wildtype 01"
# cell regions
CZ=[54,42,43,89,90,82,44,45,143,253,76,187]
P0=[545,546,566]
P1=[430,428,166,444,688] #455,
P2=[553,476,429,486,734,742,621,575,623,604]
P3=[819,824,836,851,842,830,818,848,993,1043,1044]
P4=[408,413,743,407,427,418,473,485,618,488,474,475,750,759,761,763,515,489,514,722,
527,516,774,777,787] #766,
P5=[1182,1179,1185,1163,1128,1130,1183,1153,1131,1162,1108,1109,1195,1114,1123,1115,
1073,1074,1110,1120,1116,1121,1122,1088,1089,1136,1150,1164,1146,1142,1176,1161]
P6=[958,840,953,976,977,949,961,901,935,1000,899,875,871,886,887,925,924,932,804,
762,776,773,736,817,984,775,964,933,926,923,936,832,946,922,943,967,979,815,816]
cell_regions = {"CZ": 0, "P0": 0, "P1": 0,"P2": 0,"P3": 0,"P4": 0,"P5": 0,"P6": 0 }
cell_iz = { "CZ_IZ":0,"P0_IZ": 0, "P1_IZ": 0, "P2_IZ": 0, "P3_IZ": 0, "P4_IZ": 0, "P5_IZ": 0, "P6_IZ": 0}
regions2cells = {"CZ": CZ, "P0": P0, "P1": P1, "P2": P2, "P3": P3, "P4": P4, "P5": P5, "P6": P6}
# properties to set with tissue
from openalea.stse.structures.walled_tissue_const import WalledTissueConst
const=WalledTissueConst()
const.cell_properties = {}
const.cell_properties.update( cell_regions )
const.cell_properties.update( cell_iz )
const.tissue_properties ={}
const.cell_edge_properties={}
const.wv_properties={}
const.wv_edge_properties={}
#for each property the properties dictionary will be searched. if no value is
#found, the properties will be set as default if the dictionary contains a key
#with property name it can contain a dictionary inside with
#{id: modifiedProperty}
cell_properties = {}
cell_edge_properties={}
wv_properties={}
wv_edge_properties={}
def set_zones( wt, **keys ):
"""sets zones for meristem after tissue initalization
:parameters:
wt : `WalledTissue`
Tissue on which zones will be set.
"""
for i in regions2cells:
for j in regions2cells[ i ]:
wt.cell_property(j, i, 1 )
# sets in
def set_influence_zones( wt, **keys):
def f( wt, directed_cell_edge ):
return wt.directed_cell_edge_property( directed_cell_edge, "PIN")
from openalea.stse.structures.algo.walled_tissue_influence_zones import set_property_on_tissue_component
for i in regions2cells:
#print zones[ i ]
for j in regions2cells[ i ]:
set_property_on_tissue_component( wt=wt,
cell= j,
f_component=f,
tol=0.1,
property=i+"_IZ",
property_value=1.,
with_neighbors=False,
fill_gaps=False,
additional_vertices=[])
post_procedures = [set_zones, set_influence_zones]
The tissue which is loaded with this file looks in the following way: